statsmodels
pystan
linear regression
bayesian analysis
non-informative priors
A Bayesian Hello World
Using the (a) statsmodels and (b) pystan packages, this notebook reproduces simple regression results in ARM, Section 3.1 [GH07]
Dustin Lennon
September 2019
https://dlennon.org/archive/20190918_stan001
September 2019
Simple Linear Regression
Using a Bayesian Model
Simple Linear Regression Using a Bayesian Model
Simple Linear Regression Using a Bayesian Model
We replicate the results of a classical simple linear regression using a bayesian model with non-informative priors. This is effectively “Hello, World” using the PyStan package.
import matplotlib.pyplot as plt
from matplotlib.lines import Line2D as Line2D
from matplotlib.figure import Figure as Figure
import os
import numpy as np
import pandas as pd
import statsmodels.api as sm
import pystan
import arviz as az
# pystan model cache
import sys
sys.path.append(r"./cache/mc-stan")
import model_cache
Dataset
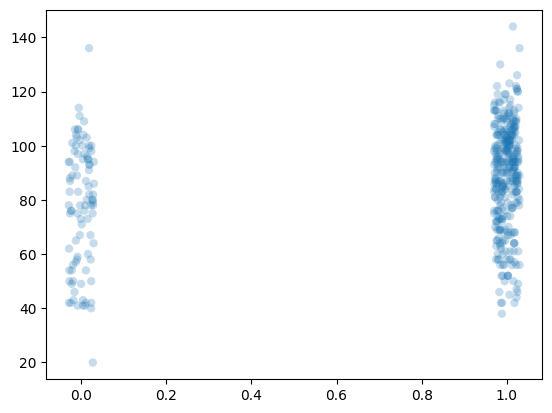
We replicate GH07, Figure 3.1. This uses the mother’s education and children’s test scores dataset.
# Load data
df = pd.read_stata("~/Workspace/Data/ARM/child.iq/kidiq.dta")
df.head()
| kid_score | mom_hs | mom_iq | mom_work | mom_age | |
|---|---|---|---|---|---|
| 0 | 65 | 1.0 | 121.117529 | 4 | 27 |
| 1 | 98 | 1.0 | 89.361882 | 4 | 25 |
| 2 | 85 | 1.0 | 115.443165 | 4 | 27 |
| 3 | 83 | 1.0 | 99.449639 | 3 | 25 |
| 4 | 115 | 1.0 | 92.745710 | 4 | 27 |
# data
x = df['mom_hs']
y = df['kid_score']
# jitter (for scatterplot)
np.random.seed(1111)
xj = x + np.random.uniform(-0.03,0.03,size=len(x))
# Scatterplot
fig = plt.figure()
ax = fig.gca()
pc = ax.scatter( *[xj,y], alpha=0.25, ec=None, lw=0)
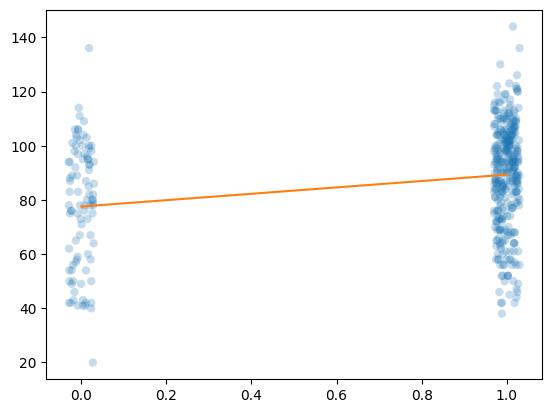
Baseline Model: Classical OLS / Simple Linear Regression
# OLS fit
X = np.column_stack(np.broadcast_arrays(1,x))
model = sm.OLS(y, X)
results = model.fit()
print(results.summary())
OLS Regression Results
==============================================================================
Dep. Variable: kid_score R-squared: 0.056
Model: OLS Adj. R-squared: 0.054
Method: Least Squares F-statistic: 25.69
Date: Fri, 06 Sep 2024 Prob (F-statistic): 5.96e-07
Time: 13:48:48 Log-Likelihood: -1911.8
No. Observations: 434 AIC: 3828.
Df Residuals: 432 BIC: 3836.
Df Model: 1
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
const 77.5484 2.059 37.670 0.000 73.502 81.595
x1 11.7713 2.322 5.069 0.000 7.207 16.336
==============================================================================
Omnibus: 11.077 Durbin-Watson: 1.464
Prob(Omnibus): 0.004 Jarque-Bera (JB): 11.316
Skew: -0.373 Prob(JB): 0.00349
Kurtosis: 2.738 Cond. No. 4.11
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
# Add a trend line based on OLS fit
yhat = results.predict([[1,0],[1,1]])
trend = Line2D([0,1], yhat, color='#ff7f0e')
ax.add_artist(trend)
ax.figure
Non-informative Bayesian Model
model_code = """
data {
int<lower=0> N;
vector[N] x;
vector[N] y;
}
parameters {
real alpha;
real beta;
real<lower=0> sigma;
}
model {
y ~ normal(alpha + beta * x, sigma);
}
"""
mod = model_cache.retrieve(model_code);
Using cached StanModel
model_data = {
'N' : len(x),
'x' : x,
'y' : y
}
fit = mod.sampling(data=model_data, iter=1000, chains=4, seed=4792)
Gradient evaluation took 4e-05 seconds
1000 transitions using 10 leapfrog steps per transition would take 0.4 seconds.
Adjust your expectations accordingly!
Iteration: 1 / 1000 [ 0%] (Warmup)
Gradient evaluation took 0.000105 seconds
1000 transitions using 10 leapfrog steps per transition would take 1.05 seconds.
Adjust your expectations accordingly!
Iteration: 1 / 1000 [ 0%] (Warmup)
Gradient evaluation took 0.000106 seconds
1000 transitions using 10 leapfrog steps per transition would take 1.06 seconds.
Adjust your expectations accordingly!
Gradient evaluation took 0.0001 seconds
1000 transitions using 10 leapfrog steps per transition would take 1 seconds.
Adjust your expectations accordingly!
Iteration: 1 / 1000 [ 0%] (Warmup)
Iteration: 1 / 1000 [ 0%] (Warmup)
Iteration: 100 / 1000 [ 10%] (Warmup)
Iteration: 100 / 1000 [ 10%] (Warmup)
Iteration: 200 / 1000 [ 20%] (Warmup)
Iteration: 100 / 1000 [ 10%] (Warmup)
Iteration: 100 / 1000 [ 10%] (Warmup)
Iteration: 300 / 1000 [ 30%] (Warmup)
Iteration: 200 / 1000 [ 20%] (Warmup)
Iteration: 200 / 1000 [ 20%] (Warmup)
Iteration: 200 / 1000 [ 20%] (Warmup)
Iteration: 400 / 1000 [ 40%] (Warmup)
Iteration: 300 / 1000 [ 30%] (Warmup)
Iteration: 300 / 1000 [ 30%] (Warmup)
Iteration: 300 / 1000 [ 30%] (Warmup)
Iteration: 400 / 1000 [ 40%] (Warmup)
Iteration: 500 / 1000 [ 50%] (Warmup)
Iteration: 501 / 1000 [ 50%] (Sampling)
Iteration: 400 / 1000 [ 40%] (Warmup)
Iteration: 400 / 1000 [ 40%] (Warmup)
Iteration: 500 / 1000 [ 50%] (Warmup)
Iteration: 501 / 1000 [ 50%] (Sampling)
Iteration: 600 / 1000 [ 60%] (Sampling)
Iteration: 500 / 1000 [ 50%] (Warmup)
Iteration: 501 / 1000 [ 50%] (Sampling)
Iteration: 500 / 1000 [ 50%] (Warmup)
Iteration: 501 / 1000 [ 50%] (Sampling)
Iteration: 700 / 1000 [ 70%] (Sampling)
Iteration: 600 / 1000 [ 60%] (Sampling)
Iteration: 600 / 1000 [ 60%] (Sampling)
Iteration: 600 / 1000 [ 60%] (Sampling)
Iteration: 700 / 1000 [ 70%] (Sampling)
Iteration: 800 / 1000 [ 80%] (Sampling)
Iteration: 700 / 1000 [ 70%] (Sampling)
Iteration: 700 / 1000 [ 70%] (Sampling)
Iteration: 800 / 1000 [ 80%] (Sampling)
Iteration: 900 / 1000 [ 90%] (Sampling)
Iteration: 800 / 1000 [ 80%] (Sampling)
Iteration: 800 / 1000 [ 80%] (Sampling)
Iteration: 900 / 1000 [ 90%] (Sampling)
Iteration: 1000 / 1000 [100%] (Sampling)
Elapsed Time: 0.120733 seconds (Warm-up)
0.070195 seconds (Sampling)
0.190928 seconds (Total)
Iteration: 900 / 1000 [ 90%] (Sampling)
Iteration: 900 / 1000 [ 90%] (Sampling)
Iteration: 1000 / 1000 [100%] (Sampling)
Elapsed Time: 0.134369 seconds (Warm-up)
0.06916 seconds (Sampling)
0.203529 seconds (Total)
Iteration: 1000 / 1000 [100%] (Sampling)
Elapsed Time: 0.139048 seconds (Warm-up)
0.07275 seconds (Sampling)
0.211798 seconds (Total)
Iteration: 1000 / 1000 [100%] (Sampling)
Elapsed Time: 0.138901 seconds (Warm-up)
0.073539 seconds (Sampling)
0.21244 seconds (Total)
print(fit)
Inference for Stan model: anon_model_6fddc89ca61cc8fa76fc51c1448e28ab.
4 chains, each with iter=1000; warmup=500; thin=1;
post-warmup draws per chain=500, total post-warmup draws=2000.
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
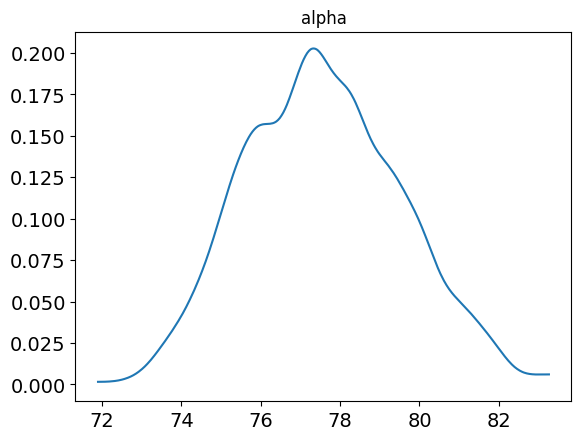
alpha 77.56 0.07 1.95 73.98 76.11 77.48 78.93 81.49 895 1.0
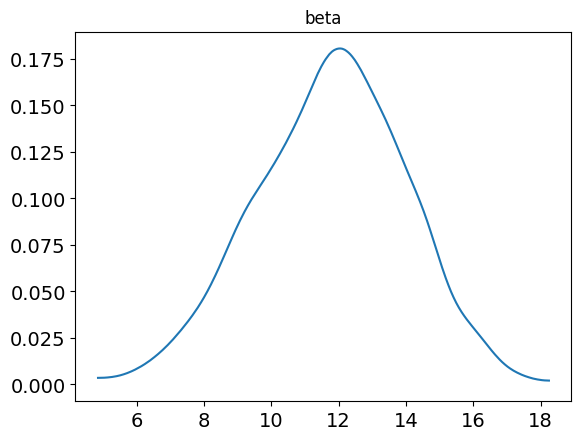
beta 11.76 0.07 2.21 7.27 10.25 11.86 13.33 15.95 954 1.0
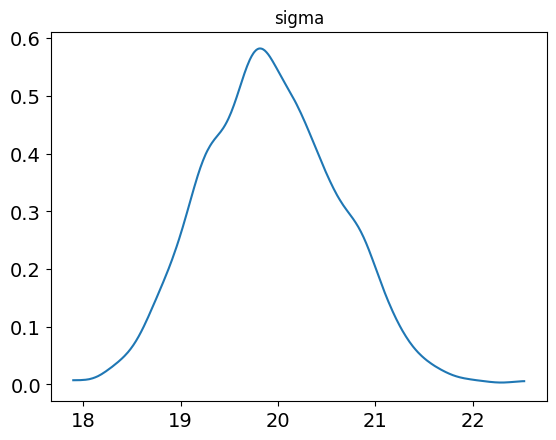
sigma 19.93 0.02 0.69 18.65 19.43 19.9 20.39 21.31 1106 1.0
lp__ -1511 0.04 1.09 -1514 -1511 -1511 -1510 -1510 750 1.0
Samples were drawn using NUTS at Fri Sep 6 13:48:49 2024.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).
Posterior Samples
samp = fit.extract(permuted=True)
def posterior_plot(samp, k):
fig = plt.gcf()
ax = fig.gca()
az.plot_kde(samp[k], ax = ax)
ax.set_title(k)
ax.figure
posterior_plot(samp, 'alpha')
posterior_plot(samp, 'beta')
posterior_plot(samp, 'sigma')
Bibliography
Bibliography
Bibliography
[GH07] Gelman, A., and Hill, J. (2007). Data Analysis Using Regression and Multilevel/Hierarchical Models. Chapter 3.
